 hlog / index-page
hlog / index-page
A tech & domain blog powered by Shtanglitza
 hlog / index-page
hlog / index-page
A tech & domain blog powered by Shtanglitza
The integration of Large Language Models (LLMs) with knowledge graphs is gaining significant traction, particularly in the context of Retrieval-Augmented Generation (RAGs). In these scenarios, LLMs usually act as interfaces for querying and summarizing information retrieved from a knowledge graph. However, other scenarios are yet to be explored. In this blog post, we explore the innovative application of LLMs for enriching structured data directly through SPARQL queries. Using the SPARQL.anything framework and the GROQ API, we'll demonstrate how to interact with a remote LLM, unlocking new possibilities for knowledge enrichment.
For those who are interested in knowledge graphs and data integration using RDF, SPARQL.anything is a powerful framework that allows users to query various data sources using the SPARQL query language. It supports querying different types of data sources, including JSON, XML, relational databases, and even remote APIs.
SPARQL.anything functions as both a CLI and a server (utilizing Apache Fuseki). For a deeper dive, you can refer to the documentation. In this experiment, we will run the server using a simple command.
java -jar sparql-anything-server-<version>.jar
The Fuseki console should be accessible at http://localhost:3000/sparql.
In our experiment, GROQ, a remote LLM API, was queried using SPARQL.anything due to its ultra-low latency and cost efficiency.
Create GROQ API keys on https://console.groq.com/keys. For these experiments, We will use mixtral-8x7b-32768.
Once the environment is set up, we can fire some SPARQL query example:
PREFIX xyz: <http://sparql.xyz/facade-x/data>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX fx: <http://sparql.xyz/facade-x/ns>
SELECT * WHERE {
SERVICE <x-sparql-anything:https://api.groq.com/openai/v1/chat/completions>
{
fx:properties fx:media-type "application/json" ;
fx:http.header.Authorization "Bearer gsk_*****" ;
fx:http.header.Content-Type "application/json" ;
fx:use-rdfs-member true ;
fx:http.method "POST" ;
fx:http.payload '{"messages": [{"role": "user", "content": "Explain the importance of fast language models"}], "model": "mixtral-8x7b-32768"}' .
?s xyz:choices/rdfs:member/xyz:message/xyz:content ?message .
}
}
The query successfully returned the requested data from the LLM GROQ API which is shown in Fuseki console.
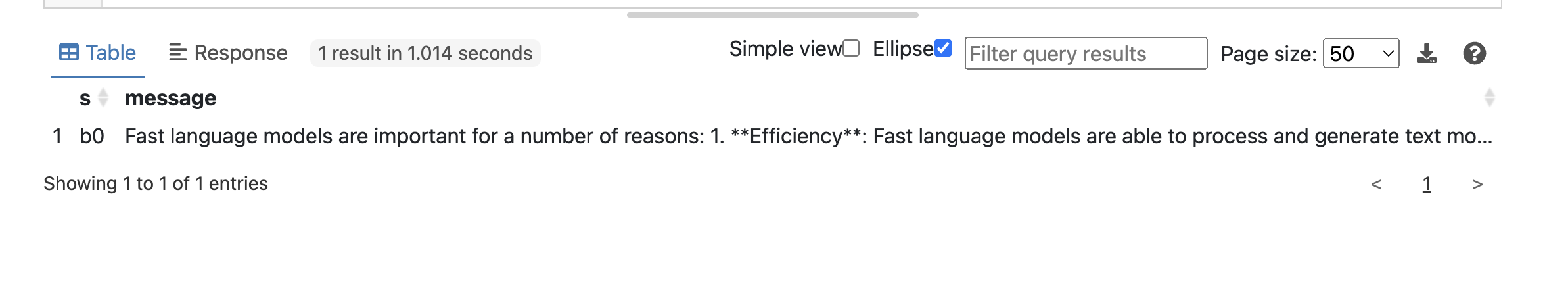
Not bad. Notice special properties such as fx:http.payload or fx:http.header.Authorization that are used to interact with the GROQ API.
Let's explore more practical use cases by applying some domain knowledge base, such as the publicly available SPARQL endpoint for Uniprot https://sparql.uniprot.org/sparql.
We can try to retrieve all genes associated with Alzheimer's disease from Uniprot and generate some preliminary hypotheses from LLM suitable for further exploration. The prompt for this task could be structured as follows:
Suggest unexplored links or genes that may need further investigation based on existing set of genes … in context of Alzheimer disease.
SPARQL Query might look like:
PREFIX xyz: <http://sparql.xyz/facade-x/data/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX fx: <http://sparql.xyz/facade-x/ns/>
PREFIX up: <http://purl.uniprot.org/core/>
PREFIX taxon: <http://purl.uniprot.org/taxonomy/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
SELECT ?genes ?payload ?message
WHERE
{
SERVICE <x-sparql-anything:https://api.groq.com/openai/v1/chat/completions> {
SERVICE <https://sparql.uniprot.org/sparql>
{
SELECT (GROUP_CONCAT(?name; separator=", ") AS ?genes)
{
?protein a up:Protein .
?protein up:organism taxon:9606 .
?protein up:encodedBy ?gene .
?gene skos:prefLabel ?name .
?protein up:annotation ?annotation .
?annotation a up:Disease_Annotation .
?annotation up:disease/skos:prefLabel ?disease .
FILTER (CONTAINS(?disease, "Alzheimer disease"))
}
}
BIND(CONCAT('{"messages": [{"role": "user", "content": "Suggest unexplored links or genes that may need further investigation based on existing set of genes: ',
coalesce(?genes),
' in context of Alzheimer disease."}], "model": "mixtral-8x7b-32768"}') AS ?payload)
fx:properties fx:media-type "application/json" ;
fx:http.header.Authorization "Bearer gsk_*****" ;
fx:http.header.Content-Type "application/json" ;
fx:use-rdfs-member true ;
fx:http.method "POST" ;
fx:http.payload ?payload.
[] xyz:choices/rdfs:member/xyz:message/xyz:content ?message .
}
}
Result:

This is useful already.
Let's investigate if we can get a more structured output from the LLM. We can apply json mode (supported from Groq API) and rephrase our prompt by introducing a system prompt for that purpose:
PREFIX xyz: <http://sparql.xyz/facade-x/data/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX fx: <http://sparql.xyz/facade-x/ns/>
SELECT ?gene
{
SERVICE <x-sparql-anything:https://api.groq.com/openai/v1/chat/completions>
{
BIND(CONCAT('{"messages": [',
'{"role": "system", "content": "You are clinical expert in domain of Alzheimer disease"},',
'{"role": "user", "content": "return most important genes linked to Alzheimer disease. Return the genes json array where every member has one field gene_name"}],',
'"model": "mixtral-8x7b-32768",',
'"temperature": 0.19,',
'"response_format": {"type": "json_object"}}') AS ?payload)
fx:properties fx:media-type "application/json" ;
fx:http.header.Authorization "Bearer gsk_*****" ;
fx:http.header.Content-Type "application/json" ;
fx:use-rdfs-member true ;
fx:http.method "POST" ;
fx:http.payload ?payload.
[] xyz:choices/rdfs:member/xyz:message/xyz:content ?content .
SERVICE <x-sparql-anything:>
{
fx:properties fx:content ?content ;
fx:media-type "application/json" .
[] fx:anySlot/xyz:gene_name ?gene .
}
}
}
returns:
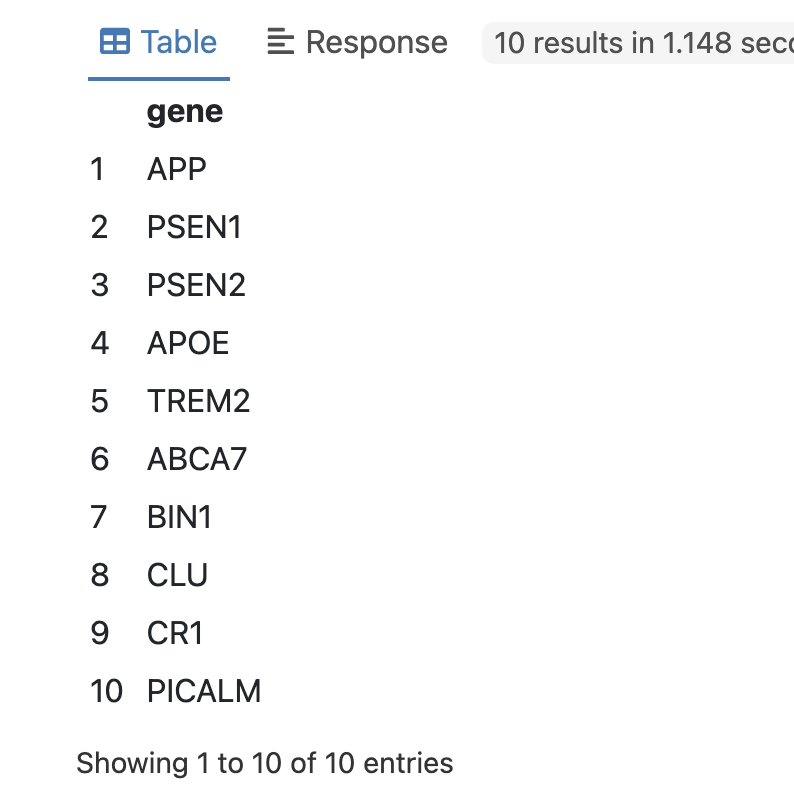
We can double-check the LLM response above by looking up the gene-disease annotation in Uniprot:
PREFIX xyz: <http://sparql.xyz/facade-x/data/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX fx: <http://sparql.xyz/facade-x/ns/>
PREFIX up: <http://purl.uniprot.org/core/>
PREFIX taxon: <http://purl.uniprot.org/taxonomy/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
SELECT ?gene ?disease
{
SERVICE <x-sparql-anything:https://api.groq.com/openai/v1/chat/completions>
{
BIND(CONCAT('{"messages": [',
'{"role": "system", "content": "You are clinical expert in domain of Alzheimer disease"},',
'{"role": "user", "content": "return most important genes linked to Alzheimer disease. Return the genes json array where every member has one field gene_name"}],',
'"model": "mixtral-8x7b-32768",',
'"temperature": 0.19,',
'"response_format": {"type": "json_object"}}') AS ?payload)
fx:properties fx:media-type "application/json" ;
fx:http.header.Authorization "Bearer gsk_******" ;
fx:http.header.Content-Type "application/json" ;
fx:use-rdfs-member true ;
fx:http.method "POST" ;
fx:http.payload ?payload.
[] xyz:choices/rdfs:member/xyz:message/xyz:content ?content .
SERVICE <x-sparql-anything:>
{
fx:properties fx:content ?content ;
fx:media-type "application/json" .
[] fx:anySlot/xyz:gene_name ?gene .
}
SERVICE <https://sparql.uniprot.org/sparql>
{
?protein a up:Protein .
?protein up:organism taxon:9606 .
?protein up:encodedBy/skos:prefLabel ?gene .
?protein up:annotation ?annotation .
?annotation a up:Disease_Annotation .
?annotation up:disease/skos:prefLabel ?disease .
}
}
}

Subsequently, the LLM GROQ API was utilized to rapidly return gene names associated with Alzheimer disease as described in prompt. Since the result from API is returned as a stringified JSON object, additional processing is done using fx:anySlot magic property. The retrieved gene names were then combined with the gene-disease annotation data obtained from the Uniprot database for further verification. This integration of data from multiple sources facilitated a comprehensive analysis of the relationship between the identified genes and their associated diseases.
What we've shown so far:
Here are some additional notes from my experiment:
fx:http.header.Authorization magic property).Published: 2024-12-25
Written by: